Efficiency of five RNA extraction protocols for Grammostola actaeon (Pocock, 1903) small spinneret tissue samples
Abstract
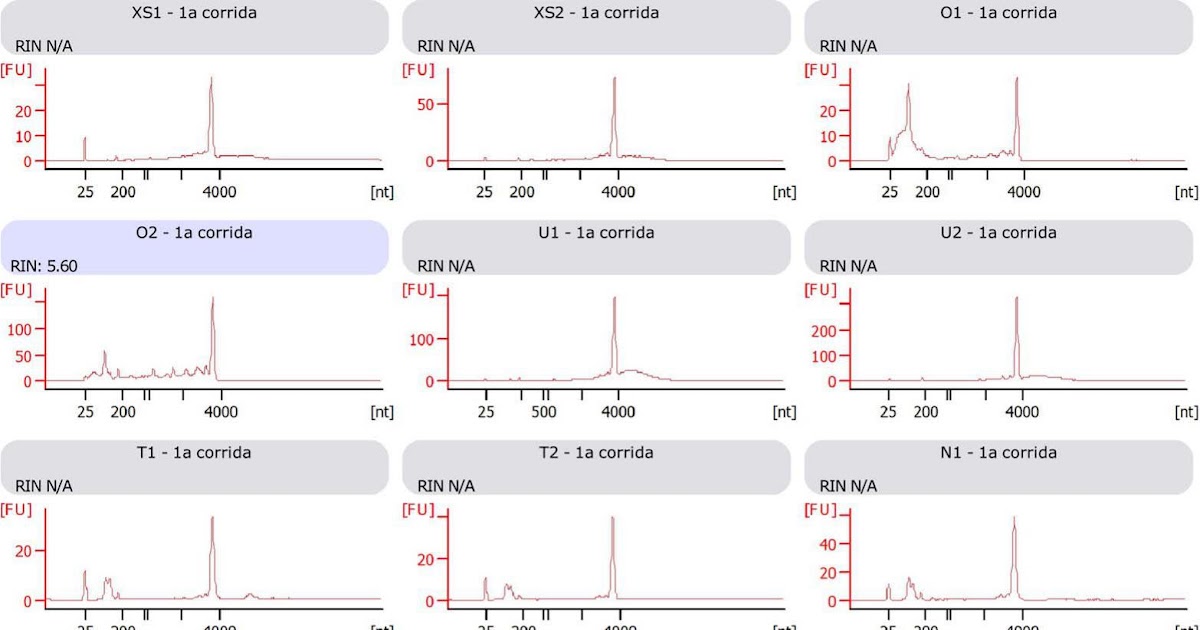
Systemic optimization of RNA extraction protocols in non-model arachnids is indispensable for gene expression studies, including transcriptome sequencing and analyses. Spinnerets of the Neotropical tarantula Grammostola actaeon (Pocock, 1903) (family Theraphosidae) were used to evaluate the performance of two RNA extraction reagents and three commercially available kits for isolating total RNA from small eukaryotic tissue samples. Total RNA was extracted from spinneret tissues, using two samples for each method. We used the commercially available reagents TRIzol and NucleoZOL and the RNA extraction kits NucleoSpin and NucleoSpin XS (Macherey Nagel) and Total RNA Purification Kit (Norgen Biotek Corp). Quantification using Qubit revealed that NucleoSpin, Norgen, and NucleoSpin XS resulted in the highest RNA yields respectively, while Nanodrop analysis ranked Norgen, TRIzol, NucleoSpin, NucleoSpin XS in descending order. Bioanalyzer analysis indicated that Nucleospin, and NucleoSpin XS delivered the best results for our samples. While each method successfully yielded sufficient RNA for RNA-seq experiments, variations in RNA quality among methods indicate differences in their suitability for specific applications. Our data provide further evidence that RNA integrity number (RIN)-based assessments in G. actaeon may not be reliable for evaluating RNA quality due to a widespread occurrence of the ‘gap deletion’ phenomenon in arthropods. RNA from species with 28S rRNA collapsed can yield high-quality transcriptomes, suggesting that current RIN-based assessments may not be reliable for evaluating RNA quality in many non-model invertebrates. Overall, differences in results of commercially available RNA extraction reagents/kits should be considered when selecting the most appropriate RNA extraction method for gene expression analysis.